Our recent publication in Bioessays, “Microbial Diversity in the Eukaryotic SAR Clade: Illuminating the Darkness Between Morphology and Molecular Data” was a collaborative effort between the Delwiche, Lane, and Katz labs. SAR taxa are members of the clade that includes Stramenopiles, Alveolates, and Rhizaria. These organisms comprise a major component of eukaryotic diversity, but because they do not include plants, animals, or fungi, many people are largely unaware of them.
The paper describes the vast gulf between the understanding of diversity in the SAR clade depending upon whether that diversity is examined with morphological or molecular methods. The problem is multifaceted: there are many organisms that have been described morphologically, but have not yet been studied with molecular methods, often because they are rare or difficult to work with in the laboratory; at the same time, environmental molecular methods have revealed a vast diversity of organisms, many of which have probably never been described morphologically, while others likely represent ‘cryptic diversity’ (i.e., organisms that are difficult to distinguish with morphological methods). Thus, there are layers of discovery to be performed in SAR taxa. We need to put more effort into describing them morphologically, into studying their sequence diversity, and into correlating these data types.
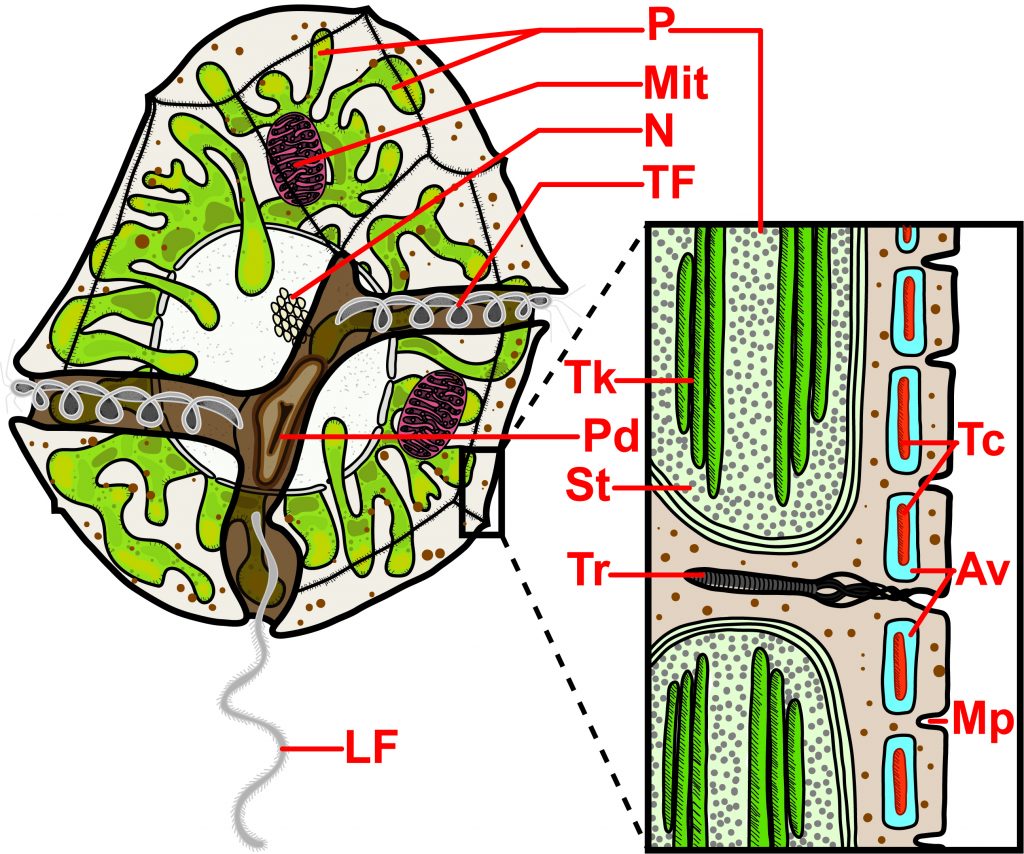
One nice feature of this paper is a figure illustrating representative species within the SAR clade. Postdoc Brittany Ott put a tremendous amount of effort into developing illustrations that accurately represent the key features of the clade and doing so with diagrams of real organisms rather than abstract representations.
Here is the direct link: http://rdcu.be/IwLB